2.5 Explanatory and response variables
The contents of this section have been extracted and modified from the article Disentangling host–microbiota complexity through hologenomics published in Nature Reviews Genetics in 2022 by the authors of the Holo-omics Workbook.
Host genomic and microbial metagenomic data generated under hologenomic setups can take on different roles when generating statistical models. While the environment is most often considered as an explanatory variable (though one can also study how the hologenome affects the environment), the host genome and the microbial metagenome are sometimes viewed as explanatory and sometimes as response variables, depending on the aim of the research. In many cases, directionality is set by the researcher rather than the biological system itself, as host-microbiota systems contain many bi-directional interactions and circular processes, which complicate the establishment of causal relationships. Here, we define three basic models in which the three main variables (genome, metagenome and environment) are assigned different roles to address different types of fundamental questions.
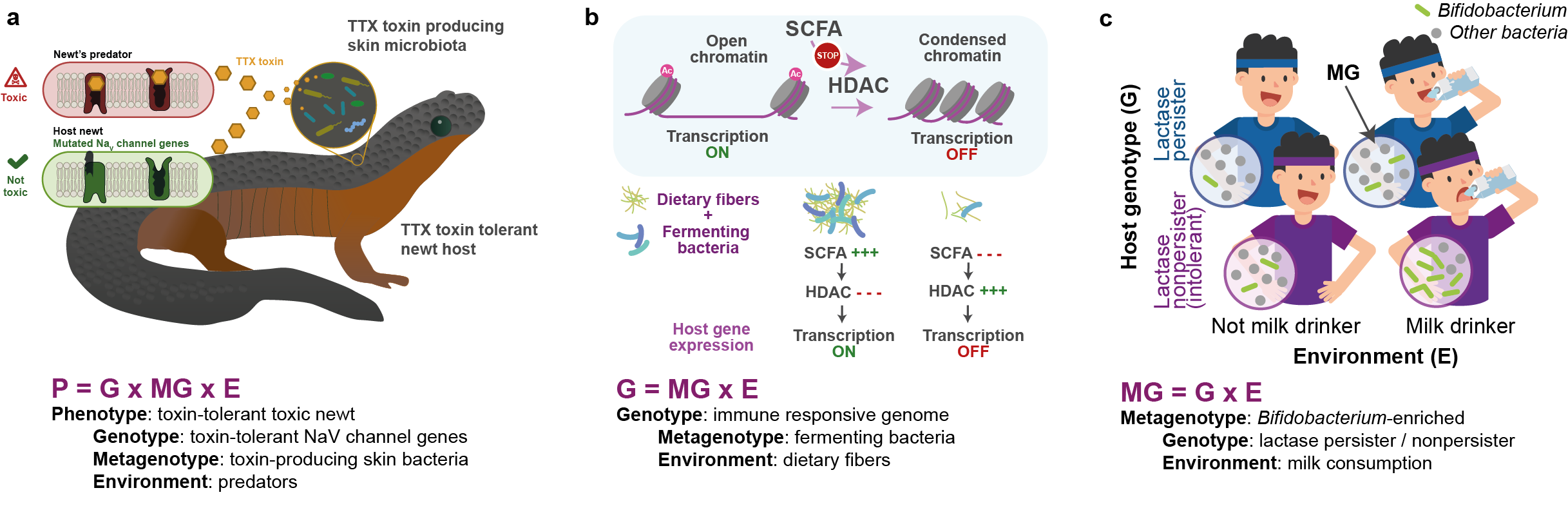
Examples of biological processes addressed by the different models of host-microbiota interactions. a) How does the hologenome shape animal phenotypes? Only the combination of specific host genomic (G) and microbial metagenomic (MG) features, probably developed due to a selective force exerted by the presence of predators (E) enables rough-skinned newts to have skin toxicity, an ecologically relevant phenotypic trait (P). b) How do the microbial metagenome and environment shape host genomic features? SCFA-producing bacteria along with a fibre-rich diet enhance chromatin accessibility and thus activate immune gene expression. c) How do the host genome and the environment shape microbial genomic features? Only the combination of a lactase nonpersister genotype combined with the milk-drinking envirotype generates a microbial metagenotype characterised by enrichment of Bifidobacterium.
Phenotype as a product of genotype, metagenotype and envirotype
This is the main model used when hologenomics is conducted to ascertain how genome-metagenome-environment interactions affect the biological properties of a host, such as disease susceptibility, performance or fitness. It is an especially common and relevant model for health, agricultural, and ecological and evolutionary research 19,125–127. One clear example of a phenotype shaped by host genomic, microbial metagenomic and environmental factors was recently reported for rough-skinned newts. The study showed that bacteria on the skin of the newts produce a deadly neurotoxin from which the newt is protected by mutations in five host genes that encode the NaV channels normally targeted by the toxin. Thus, this ‘toxic newt’ phenotype is the result of both host and microbial genes, which likely evolved under the pressure exerted by an environmental factor, namely the presence of predators.
Genotype expression influenced by metagenotype and envirotype
When studying how core host genomic features, which contribute to shaping phenotypes, are affected by the microbiota, host genomic features become the response variable. Unlike the microbial metagenome, the genome sequence of the host organism is not variable, but microorganisms can induce chromatin remodelling and DNA methylation, and thus modulate the bioactivity of molecular receptors and host gene expression. A well-studied pathway that links the microbiota with host gene expression involves modulation of the activity of host histone deacetylases (HDAC) by short chain fatty acids (SCFA) produced by intestinal microorganisms. HDACs remove histone lysine acetyl groups, which leads to chromatin condensation and transcriptional silencing of genes. Increased SCFA concentrations inhibit histone deacetylases, thereby enhancing chromatin accessibility and activating gene expression. A metagenotype with a higher capacity to produce SCFAs combined with an envirotype characterised as a fibre-rich diet (required to produce SCFAs), therefore contributes to boost immune response through activating host immune gene expression.
Metagenotype as a product of genotype and envirotype
This model assumes the inverse causal directionality between the host genome and microbial metagenome to that described above. Candidate host genes related to microbiota features can be identified through GWAS in which the metagenotype (or derived metrics such as diversity or abundance of specific microbial taxa, genes or metabolic functions) are treated as a phenotypic trait. For instance, the increased abundance of lactose degrader Bifidobacteria in humans has been shown to be associated with lactase nonpersister genotype and consumption of milk (envirotype). Once candidate genes are known, targeted analyses in which natural or human-controlled genomic variability (such as the number of copies of the amylase-encoding gene in humans) can be contrasted under controlled environmental conditions to ascertain the effect on metagenotypes (such as the abundance of Ruminococcaceae bacteria in the gut microbiota).