1.1 Omic layers
Nucleic acid sequencing and mass spectrometry technologies that enable tracking the biomolecular pathways linking host and microbial genomic sequences with biomolecular phenotypes by generating (meta)transcriptomes, (meta) proteomes, and (meta)metabolomes. The same technologies also enable epigenomic and exposomic profiling, which can further contribute to disentangling the biochemical associations between host-microbiota-environment interactions and their effect on host phenotypes.
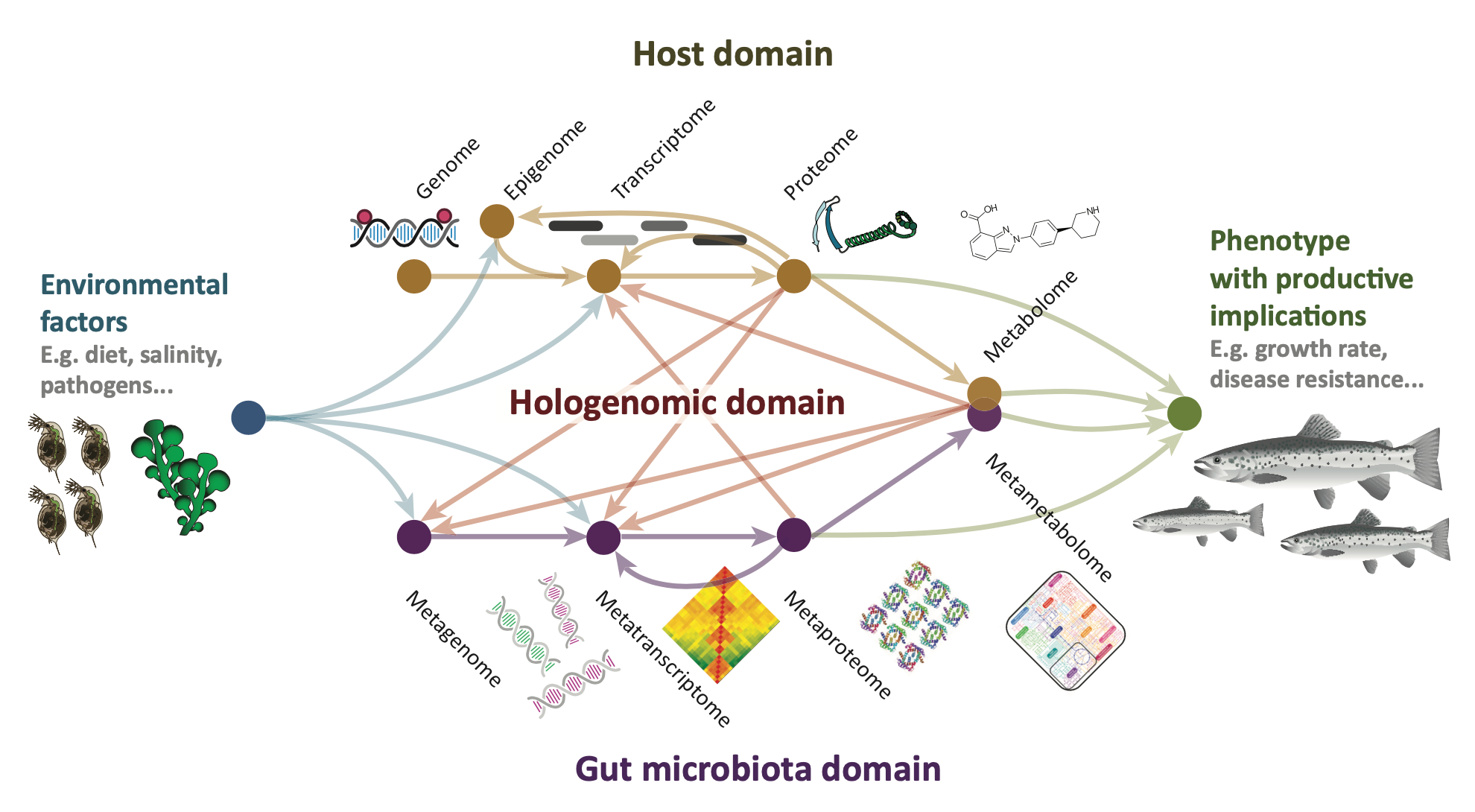
Overview of omic layers. Modified from Limborg et al. 2018 [2].
In this workbook we consider seven omic layers that require specific data generation and analysis strategies before integrating them into multi-omic statistical models:
- Nucleic acid sequencing-based
- Mass spectrometry-based
Acknowledging the distinct biological and structural characteristics of these seven omic layers is essential to design experiments and analytical pipelines for better solving the complex puzzle of host-microbiota interactions.
Host genomics (HG)
Host genomics refers to the study of an organism’s genetic information (its genome) and how it relates to that organism’s traits and characteristics. In the case of host genomics, we are specifically looking at the genetic information of a host organism, such as a human or an animal, and how that genetic information can influence things like susceptibility to certain diseases, response to treatments, and traits related to its microbiome composition and function.
Host transcriptomics (HT)
Host transcriptomics refers to the study of the full set of RNA molecules produced by an organism’s cells (known as the transcriptome), and how it relates to that organism’s traits and characteristics. In the case of host transcriptomics, we are specifically looking at the RNA molecules produced by the cells of a host organism, such as a human or an animal, and how those RNA molecules can influence things like gene expression, protein production, and how this affects the host’s associated microbiome.
Microbial metagenomics (MG)
Microbial metagenomics is the study of genetic material from a mixed community of microorganisms, without the need to isolate and culture individual organisms. This approach involves extracting DNA from an environmental sample, including e.g. from the intestinal environment of a host organism, and sequencing it to obtain a snapshot of the genetic diversity and potential functions of the microbial community present in that sample. By analysing the genetic information obtained through metagenomics, researchers can gain insights into the metabolic capabilities, ecological roles, and evolutionary relationships of the microorganisms living in a particular environment.
Microbial metatranscriptomics (MT)
Microbial metatranscriptomics is the study of all the genetic material expressed as RNA transcripts by a community of microorganisms living in a particular environment. This approach allows researchers to understand which genes are active and which metabolic pathways are being used by the microorganisms in a specific ecosystem. Essentially, it involves analyzing the RNA molecules produced by the microorganisms in a sample to gain insight into their activities and behaviors.
Host proteomics (HP)
Host proteomics is the study of all the proteins produced by a specific host organism in response to various stimuli, including disease or infection. Proteins are the workhorses of the body and perform many important functions, such as regulating cell growth, repairing damaged tissues, and fighting infections. Proteomics techniques involve analysing the entire set of proteins, or proteome, of a particular organism or tissue sample, to understand how they are produced, modified, and interact with each other. The most popular method to generate proteomics data today is mass spectrometry which involves ionizing protein samples and analyzing the resulting ions based on their mass-to-charge ratio to identify the protein and its modifications.
Microbial metaproteomics (MP)
Microbial metaproteomics is the large-scale study of all the proteins produced by a community of microorganisms living in a particular environment such as a host organism. This approach involves extracting and analysing proteins from a mixed microbial sample, without the need to isolate and culture individual microorganisms. By identifying and quantifying the proteins present in the sample, microbial metaproteomics can provide insights into the functional roles and metabolic activities of the microorganisms in the community, as well as their interactions with each other and with their environment. These data are also often generated using mass spectrometry methods as described for host proteomics above.
(Meta)metabolomics (ME)
Host as well as Meta-metabolomics is the study of all the small molecules, or metabolites, produced by an organism and/or microbial community under different physiological conditions. These metabolites include molecules such as sugars, amino acids, and lipids, which are the building blocks and energy sources for cells. Metabolomics techniques involve the identification and quantification of these molecules using advanced analytical methods, such as mass spectrometry. By analysing the metabolic profile of an organism and/or microbial community, researchers can gain insights into the biochemical pathways and metabolic networks that regulate various physiological processes in e.g. the intestinal environment of animals.