2.1 Hologenomic complexity
The contents of this section have been extracted and modified from the article Disentangling host–microbiota complexity through hologenomics published in Nature Reviews Genetics in 2022 by the authors of the Holo-omics Workbook.
Hologenomic complexity can be broadly defined as the amount of information relevant to the study that the biological system under analysis contains and it can be decomposed into three major elements: host genomic, microbial metagenomic and environmental complexity [7]. Within each of these elements, two sources of complexity can be defined: the intrinsic complexity of the system under study, including host genome size and number of bacterial genomes, and the complexity introduced by the degree of difference between the organisms under comparison such as gene expression differences versus distinct genomes.
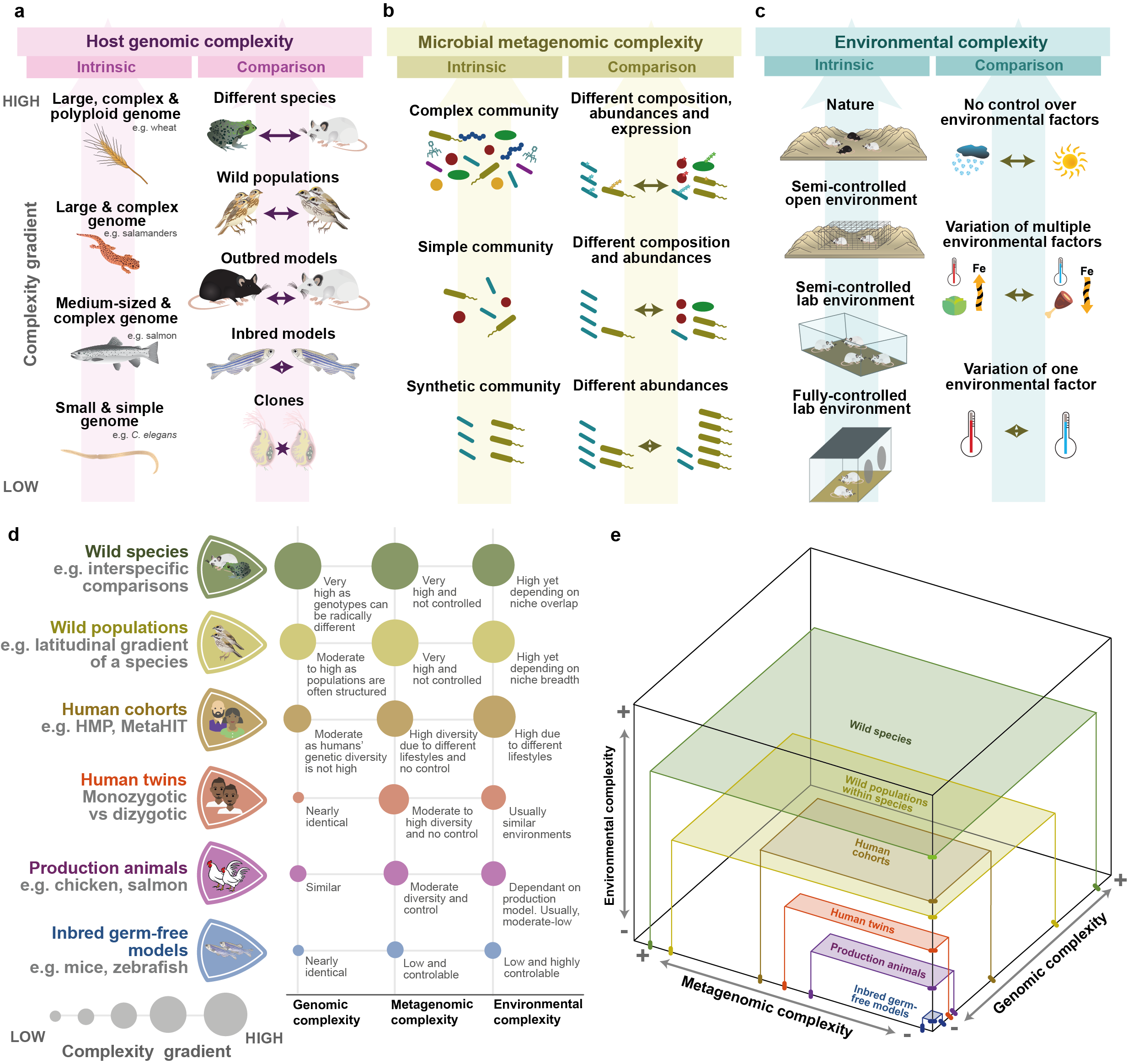
Decomposition of hologenomic complexity. (a-c) The design and interpretation of hologenomic studies depend on the host genomic (part a), microbial metagenomic (part b) and environmental (part c) complexity of the system under study. Within each axis of complexity, two types of gradients can be defined based on whether the features are intrinsic to the system or introduced by the researcher through the selection of groups under comparison. (d) Six examples of study systems with different levels of genomic, metagenomic and environmental complexity. (e) Three-dimensional representation of the complexity of the examples. The area of the plain represents the combined host genomic and microbial metagenomic complexity of the system, while the height represents the environmental complexity. The combined three-dimensional volume represents the overall hologenomic complexity of the system. HMP: Human Microbiome Project.